You are looking for information, articles, knowledge about the topic nail salons open on sunday near me q5 site directed mutagenesis kit on Google, you do not find the information you need! Here are the best content compiled and compiled by the https://chewathai27.com/to team, along with other related topics such as: q5 site directed mutagenesis kit q5 site-directed mutagenesis kit (without competent cells), q5® site-directed mutagenesis kit pdf, q5® site-directed mutagenesis kit price, q5 site-directed mutagenesis kit protocol, site-directed mutagenesis kit neb, q5 site-directed mutagenesis troubleshooting, quikchange site-directed mutagenesis kit, agilent site-directed mutagenesis
What is the best site-directed mutagenesis kit?
QuikChange Site-Directed Mutagenesis Kit, from Stratagene – is the best one.
How do you use Neb base changer?
- Enter the starting plasmid sequence.
- Choose your mutagenesis experiment.
- Define the mutation region.
- [Enter Desired Sequence] (not required for deletions)
- View primers and annealing temperature.
How do you design primers for site-directed mutagenesis?
…
How do I insert a specific sequence into any vector?
- add the vector sequence of your choice.
- choose the insertion locus (independent of restriction sites) and specify the exact nucleotides to be added.
- download primer and PCR information based on your design.
What is Quick Change PCR?
The PCR Quick Change or site directed mutagenesis is used to change DNA bases on a sequence of interest (maximum 5 bases). The most important step in this experiment is the design of the primers.
What is QuikChange directed mutagenesis?
The QuikChange Multi site-directed mutagenesis system is a novel technology that allows mutagenesis at multiple sites in a single round, using a single oligonucleotide per site. This system simplifies randomizing key amino acids using oligos containing degenerate codons.
Which polymerase is used in site-directed mutagenesis?
During the study we found that the Taq DNA polymerase used for PCR adds on a single extra base (usually an A) at the end of a large fraction of the newly synthesized chains. These had to be removed by the Klenow fragment of DNA polymerase to insure restoration of the gene sequence.
Is Crispr site-directed mutagenesis?
CRISPR/Cas9 has emerged as a rapidly programmable molecular tool for targeting specific sequences of DNA, thus providing a mechanism for markerless selection when performing site-directed mutagenesis.
What are the four different types of PCR based site-directed mutagenesis?
- Figure 1. Site-directed mutagenesis by traditional PCR. Primers incorporating the desired base changes are used in PCR. …
- Figure 2. Site-directed mutagenesis by primer extension. …
- Figure 3. Site-directed mutagenesis by inverse PCR.
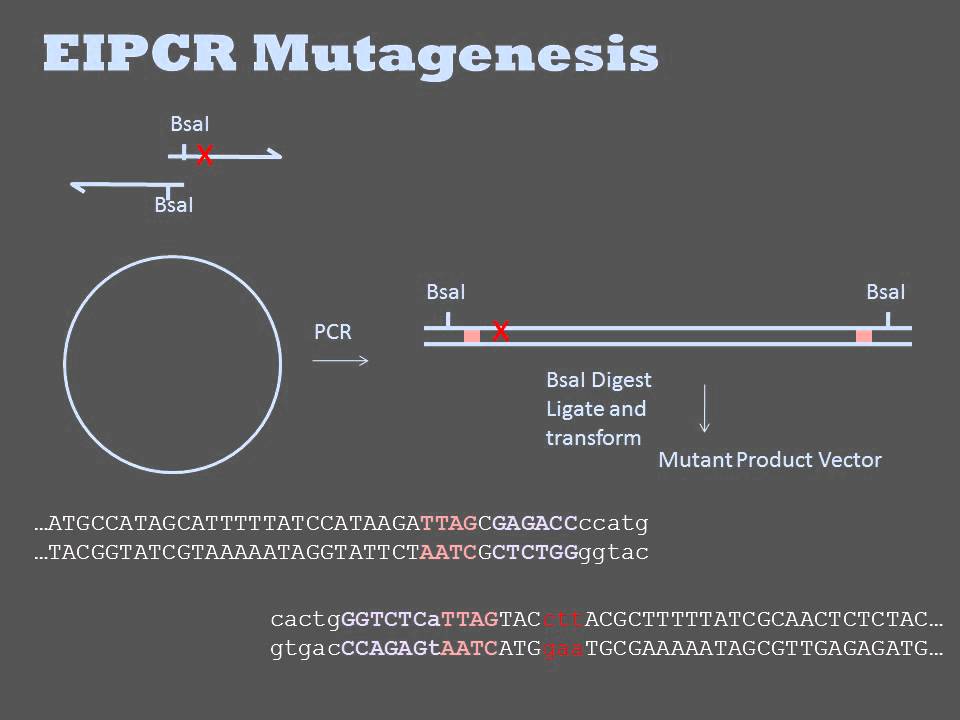
What are back to back primers?
The back-to-back primer design allows for deletions of unlimited size to be generated simply by positioning both 5′ ends of forward and reverse primers directly on the sequence flanking the desired deletion.
What is the purpose of KLD treatment?
KLD Enzyme Mix is a unique blend of Kinase, Ligase and DpnI enzymes. This formulation allows efficient phosphorylation, intramolecular ligation/circularization and template removal in a single 5 minute reaction step at room temperature.
How do you design a primer?
- Aim for the GC content to be between 40 and 60% with the 3′ of a primer ending in G or C to promote binding. …
- A good length for PCR primers is generally around 18-30 bases. …
- Try to make the melting temperature (Tm) of the primers between 65°C and 75°C, and within 5°C of each other.
What is forward primer and reverse primer?
The forward primer attaches to the start codon of the template DNA (the anti-sense strand), while the reverse primer attaches to the stop codon of the complementary strand of DNA (the sense strand). The 5′ ends of both primers bind to the 3′ end of each DNA strand.
How do you make a mutant primer?
Primers should be between 25 and 45 bases in length, with a melting temperature (Tm) of ≥78°C. The desired mutation (deletion or insertion) should be in the middle of the primer with ~10–15 bases of correct sequence on both sides and minimum GC content of 40% and should terminate in one or more C or G bases.
What is SDM protocol?
SDM is an in vitro procedure that uses custom designed oligonucleotide primers to confer a desired mutation in a double-stranded DNA plasmid. Formerly, a method pioneered by Kunkel (Kunkel, 1985) that takes advantage of a strain deficient in dUTPase and uracil deglycosylase so that the recipient E.
New England Biolabs (UK) Ltd – Q5® Site-Directed Mutagenesis Kit
- Article author: www.neb.uk.com
- Reviews from users: 40967
Ratings
- Top rated: 3.7
- Lowest rated: 1
- Summary of article content: Articles about
New England Biolabs (UK) Ltd – Q5® Site-Directed Mutagenesis Kit
The Q5 Site-Directed Mutagenesis Kit enables rap, site-specific mutagenesis of double-stranded plasm DNA in less than 2 hours (Figure 1). The kit utilizes … … - Most searched keywords: Whether you are looking for
New England Biolabs (UK) Ltd – Q5® Site-Directed Mutagenesis Kit
The Q5 Site-Directed Mutagenesis Kit enables rap, site-specific mutagenesis of double-stranded plasm DNA in less than 2 hours (Figure 1). The kit utilizes … Q5® Site-Directed Mutagenesis Kitgene art;GeneArt;gene-art;quick change;QuickChange;quick-change - Table of Contents:
 Read More
Read More
Site-directed Mutagenesis – YouTube
- Article author: www.youtube.com
- Reviews from users: 36663
Ratings
- Top rated: 4.9
- Lowest rated: 1
- Summary of article content: Articles about Site-directed Mutagenesis – YouTube Updating …
- Most searched keywords: Whether you are looking for Site-directed Mutagenesis – YouTube Updating Site-directed mutagenesis involves making localized edits to a preexisting DNA sequence. Typically it is used to introduce point mutations into a sequence s…video, chia sẻ, điện thoại có máy ảnh, điện thoại quay video, miễn phí, tải lên
- Table of Contents:
NEBaseChanger
- Article author: nebasechanger.neb.com
- Reviews from users: 26171
Ratings
- Top rated: 3.5
- Lowest rated: 1
- Summary of article content: Articles about NEBaseChanger Updating …
- Most searched keywords: Whether you are looking for NEBaseChanger Updating NEBaseChanger can design primers specific to the mutagenesis experiment you are performing and calculate a recommended custom annealing temperature.
- Table of Contents:

Insertions, deletions, and substitutions, oh my!—designing primers for site-directed mutagenesis
- Article author: www.takarabio.com
- Reviews from users: 33752
Ratings
- Top rated: 3.8
- Lowest rated: 1
- Summary of article content: Articles about Insertions, deletions, and substitutions, oh my!—designing primers for site-directed mutagenesis Updating …
- Most searched keywords: Whether you are looking for Insertions, deletions, and substitutions, oh my!—designing primers for site-directed mutagenesis Updating Design primers for generating deletions, base substitutions, or additions using a flexible, accurate system.in fusion cloning, PCR cloning, ligation independent cloning, site directed mutagenesis kit,
PCR based site directed mutagenesis, PCR cloning primer design - Table of Contents:
What is the protocol for In-Fusion Cloning site-directed mutagenesis
How do I design In-Fusion primers for site-directed mutagenesis
Deleting a specific sequence from any vector
Inserting a specific sequence in any vector
Deleting and replacing a sequence in any vector
Partner with Takara Bio!
Log in to enjoy additional benefits
Want to save this information
That’s GOOD Science!

Q5® Site Directed Mutagenesis Kit – New England Biolabs
- Article author: www.neb-online.fr
- Reviews from users: 24423
Ratings
- Top rated: 4.2
- Lowest rated: 1
- Summary of article content: Articles about Q5® Site Directed Mutagenesis Kit – New England Biolabs Ce kit est conçu pour la réalisation rape et efficace d’insertions, de délétions et de substitutions au niveau d’un ADN plasmique double brin. Première … …
- Most searched keywords: Whether you are looking for Q5® Site Directed Mutagenesis Kit – New England Biolabs Ce kit est conçu pour la réalisation rape et efficace d’insertions, de délétions et de substitutions au niveau d’un ADN plasmique double brin. Première …
- Table of Contents:
Principe
Liste des produits disponibles
Comparaison avec un produit concurrent

Q5 Site-Directed Mutagenesis Kit – New England Biolabs GmbH
- Article author: www.neb-online.de
- Reviews from users: 34410
Ratings
- Top rated: 3.4
- Lowest rated: 1
- Summary of article content: Articles about Q5 Site-Directed Mutagenesis Kit – New England Biolabs GmbH Das Q5® Site-Directed Mutagenesis Kit wurde entwickelt um schnell und effizient Insertionen, Deletionen und Substitutionen in Doppelstrang-DNA einzuführen. Im … …
- Most searched keywords: Whether you are looking for Q5 Site-Directed Mutagenesis Kit – New England Biolabs GmbH Das Q5® Site-Directed Mutagenesis Kit wurde entwickelt um schnell und effizient Insertionen, Deletionen und Substitutionen in Doppelstrang-DNA einzuführen. Im … Das NEB Q5® Site-Directed Mutagenesis Kit ermöglicht Ihnen die gerichtete Mutagenese von den verschiedensten Ausgangsplasmiden in nur 2 Stunden.
- Table of Contents:
Methode
Produkttabelle
Vergleich mit anderen Methoden

New England Biolabs, Inc. Q5 Site-Directed Mutagenesis Kit – 10 reactions
| Fisher Scientific
- Article author: www.fishersci.com
- Reviews from users: 42396
Ratings
- Top rated: 4.2
- Lowest rated: 1
- Summary of article content: Articles about
New England Biolabs, Inc. Q5 Site-Directed Mutagenesis Kit – 10 reactions
| Fisher Scientific The Q5 Site-Directed Mutagenesis Kit enables rap, site-specific mutagenesis of double-stranded plasm DNA in less than 2 hours. The kit utilizes the … … - Most searched keywords: Whether you are looking for
New England Biolabs, Inc. Q5 Site-Directed Mutagenesis Kit – 10 reactions
| Fisher Scientific The Q5 Site-Directed Mutagenesis Kit enables rap, site-specific mutagenesis of double-stranded plasm DNA in less than 2 hours. The kit utilizes the … New England Biolabs, Inc. Q5 Site-Directed Mutagenesis Kit – 10 reactionsShop New England Biolabs, Inc. Q5 Site-Directed Mutagenesis Kit – 10 reactions at Fishersci.com - Table of Contents:

| Fisher Scientific
Q5® Site-Directed Mutagenesis (E0554)
- Article author: www.protocols.io
- Reviews from users: 34953
Ratings
- Top rated: 4.9
- Lowest rated: 1
- Summary of article content: Articles about Q5® Site-Directed Mutagenesis (E0554)
Q5® Site-Directed Mutagenesis (E0554) V.1 … Assemble the following reagents in a thin-walled PCR tube. … FINAL CONC. … Mix reagents completely. … - Most searched keywords: Whether you are looking for Q5® Site-Directed Mutagenesis (E0554)
Q5® Site-Directed Mutagenesis (E0554) V.1 … Assemble the following reagents in a thin-walled PCR tube. … FINAL CONC. … Mix reagents completely. This is the protocol for the Q5® Site-Directed Mutagenesis Kit (E0554) - Table of Contents:

See more articles in the same category here: Top 455 tips update new.
Q5® Site-Directed Mutagenesis Kit
The Q5 Site-Directed Mutagenesis Kit enables rapid, site-specific mutagenesis of double-stranded plasmid DNA in less than 2 hours (Figure 1). The kit utilizes the robust Q5 Hot Start High-Fidelity DNA Polymerase along with custom mutagenic primers to create insertions, deletions and substitutions in a wide variety of plasmids. After PCR, the amplified material is added directly to a unique Kinase-Ligase-DpnI (KLD) enzyme mix for rapid (5 minutes), room temperature circularization and template removal (Figure 2). Transformation into high-efficiency NEB 5-alpha Competent E. coli, provided with the kit, ensures robust results with plasmids up to at least 20 kb in length.
Figure 1: Site-specific mutagenesis proceeds in less than 2 hours.
The use of a master mix, a unique multi-enzyme KLD enzyme mix, and a fast polymerase ensures that, for most plasmids, the mutagenesis reaction is complete in less than two hours.
This kit is designed for rapid and efficient incorporation of insertions, deletions and substitutions into doublestranded plasmid DNA. The first step is an exponential amplification using standard primers and a master mix fomulation of Q5 Hot Start High-Fidelity DNA Polymerase. The second step involves incubation with a unique enzyme mix containing a kinase, a ligase and DpnI. Together, these enzymes allow for rapid circularization of the PCR product and removal of the template DNA. The last step is a high-efficiency transformation into chemicallycompetent cells (provided).Figure 3: Primer Design for the Q5 Site-Directed Mutagenesis Kit
Substitutions, deletions and insertions are incorporated into plasmid DNA through the use of specifically designed forward (black) and reverse (red) primers. Unlike kits that rely on linear amplification, primers designed for the Q5 Site-Directed Mutagenesis Kit should not overlap to ensure that the benefits of
exponential amplification are realized. A) Substitutions are created by incorporating the desired nucleotide change(s) (denoted by *) in the center of the forward primer, including at least 10 complementary nucleotides on the 3´side of the mutation(s). The reverse primer is designed so that the 5´ ends of the two primers anneal back-to- back. B) Deletions are engineered by designing standard, non-mutagenic forward and reverse primers that flank the region to be deleted. C) Insertions less than or equal to 6 nucleotides are incorporated into the 5´ end of the forward primer while the reverse primer anneals back-to-back with the 5´ end of the complementary region of the forward primer. D) Larger insertions can be created by incorporating half of the desired insertion into the 5´ ends of both primers. The maximum size of the insertion is largely dictated by oligonucleotide synthesis limitations. Figure 4: NEB’s Q5 SDM Kit delivers higher transformation efficiency than Agilent’s QuikChange® SDM Kit
Results from a substitution reaction (4 nt) using the back-to-back Control SDM Primer Mix and Control SDM Plasmid (6.7 kb) are shown, along with results from a 12 nt deletion experiment (5.8 kb plasmid) and an 18 nt insertion experiment (7.0 kb plasmid). In all three cases, over 90% of the resultant colonies had incorporated the desired mutation(s). Results are normalized to total transformants if cells were not diluted prior to plating. For comparison, the same substitution reaction (4 nt) was performed with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) following Agilent’s protocol and using Agilent’s primer design tool to design overlapping primers.
*Note that the QuikChange kit does not accommodate deletions and insertions of this size, so no comparison could be made for these experiments.
Kit Components
The following reagents are supplied with this product:
NEB # Component Name Component # Stored at (°C) Amount Concentration E0554S Multi-temperature Q5® Hot Start High-Fidelity 2X Master Mix M0494AVIAL -20 1 x 0.125 ml 2 X KLD Reaction Buffer B0554AVIAL -20 1 x 0.15 ml 2 X KLD Enzyme Mix M0554AVIAL -20 1 x 0.01 ml 10 X Control SDM Primer Mix S0554AVIAL -20 1 x 0.01 ml 10 µM Control SDM Plasmid N0554AVIAL -20 1 x 0.01 ml 5 µg/ml pUC19 Vector N3041AVIAL -20 1 x 0.025 ml 50 pg/µl SOC Outgrowth Medium B9020SVIAL 4 1 x 25 ml Not Applicable NEB® 5-alpha Competent E. coli (High Efficiency) C2987HVIAL -80 10 x 0.05 ml Not Applicable
Advantages and Features
Features Generation of mutations, insertions or deletions in plasmid DNA
Non-overlapping primer design ensures robust, exponential amplification, generating a high percentage of desired mutations from a wide range of templates
Intramolecular ligation and transformation into NEB high-efficiency competent cells results in high colony yield
Extremely low error rate of Q5 Hot Start High-Fidelity DNA Polymerase reduces screening time
Hot start polymerase enables room temperature reaction set-up
DpnI background reduction permits a wide range of starting template concentrations
Use of standard primers eliminates additional expenses from phosphorylated or purified oligos
Easy-to-use PCR master mix and unique multi-enzyme KLD mix offer convenience and quality
Rapid and direct treatment step proceeds at room temperature in 5 minutes
Properties & Usage
Related Products
Product Notes
Storage Note:
The Q5 Site-Directed Mutagenesis Kit is stable at -80°C for one year. For convenience, the Q5 Hot Start High-Fidelity 2X Master Mix, KLD Enzyme Mix, KLD Reaction Buffer, Control Primers and Template DNA are packaged together in a separate box that can be removed and stored at -20°C for two years with no loss of activity. The SOC can be removed and stored at room temperature. It is important to store the NEB 5-alpha Competent E. coli at -80°C, and avoid repeated freeze-thaw cycles.
References
Insertions, deletions, and substitutions, oh my!—designing primers for site-directed mutagenesis
Does your mutagenesis method perform poorly with GC-rich templates? Are you getting too few or too many colonies, colonies without the mutation you want, or no colonies at all? Sadly, these problems have been frustrating researchers for decades.
You’ll be happy to know that there is a solution, and it starts and ends with “In-Fusion.” In-Fusion is well-known for being the highest performing seamless cloning product on the market, but did you know that you can also use the technology for site-directed mutagenesis?
In-Fusion Cloning products provide the flexibility to generate single or multiple base changes, deletions, and insertions using any vector, with over 95% accuracy—by combining the power of In-Fusion Cloning technology with high-fidelity inverse PCR using a polymerase that works well with GC-rich templates.
What is the protocol for In-Fusion Cloning site-directed mutagenesis?
During inverse PCR with In-Fusion systems, primers are oriented in opposite directions on your circular cloning vector (Figure 1). To perform mutagenesis, design your PCR primers so that they have a 15-bp overlap with each other at their 5′ ends and incorporate the mutation of interest, and use a high-fidelity PCR polymerase such as PrimeSTAR Max DNA Polymerase, which exhibits minimal error rates on GC-rich templates.
Figure 1. Procedure for performing mutagenesis with In-Fusion technology. The area where mutagenesis occurs is shown in yellow. After designing your experiment, perform the protocol (Step 3 above) on Day 1, and recover your final construct (Step 4 above) on Day 2. Note: Although all examples shown here involve protein coding (gene) sequences, you can use the same methods to modify noncoding sequences such as promoters or transcription factors.
Want the nitty-gritty details? See our Mutagenesis with In Fusion Cloning tech note!
How do I design In-Fusion primers for site-directed mutagenesis?
Our online primer design tool lets you design primers for your site-directed mutagenesis experiment simply by inputting the DNA sequences of your vector and insert(s). It allows you to specify the exact nucleotides to be deleted, add the vector sequence of your choice at your desired insertion locus, enter a replacement sequence (independent of restriction sites), and download primer and PCR information based on your design. Use our tool together with the guidelines below, noting that specialized tutorials are available for designing primers to generate deletions, insertions, and substitutions.
General guidelines for primer design
Each PCR primer should direct DNA synthesis in the opposite orientation of the other on a circular vector template.
The 3′ ends of the forward and reverse PCR primers should have 18–25 nt that are complementary to the template, ensuring efficient and specific amplification.
Mutations should be incorporated within the homologous 15-nt overlap located at the 5′ ends of the forward and reverse PCR primers (this homologous overlap is required for the recircularization of the mutated vector). Single- or multiple-base changes, deletions, or insertions can be introduced in a single In‑Fusion reaction. A larger deletion of any desirable length can also be introduced by positioning the 3′ ends of the forward and reverse primers at the border sites of a deletion, with homologous overhangs carried by the 5′ end of either of the primers.
The resulting inverse PCR will generate a linear double-stranded vector with the flanking ends complementary to each other and carrying the 15-nt homologous overlap. This overlap will be joined through the In‑Fusion reaction and recovered in E. coli, thus generating a mutated vector. Vectors amplified by inverse PCR may be treated with Cloning Enhancer to destroy the parental vector and used directly in the In-Fusion reaction.
How do I delete a specific sequence from any vector?
Use the primer design tool (selecting “Mutagenesis” under “Project Type”) together with the “Deleting a sequence” tutorial to design PCR primers to:
delete a specific base or sequence in the vector of choice
specify the exact nucleotide(s) to be deleted
generate and download the designed primers and PCR information
How do I insert a specific sequence into any vector?
Simply use the tool with the “Inserting a sequence” tutorial to design primers to:
add the vector sequence of your choice
choose the insertion locus (independent of restriction sites) and specify the exact nucleotides to be added
download primer and PCR information based on your design
How do I delete and replace a sequence in any vector?
Once again, use the tool with the “Deleting and replacing a sequence in any vector” tutorial to design primers to:
add the vector sequence of your choice
specify the exact nucleotides to be deleted
enter your replacement sequence
download primer and PCR information based on your design
With In-Fusion for mutagenesis, you now have the courage and confidence to overcome your site-directed mutagenesis challenges, and you’ll never look back with this powerful, precise tool for faster, easier, and more accurate site-directed mutagenesis using your vector of choice.
Q5® Site Directed Mutagenesis Kit
La mutagenèse dirigée consiste à introduire des mutations (insertions, délétions et substitutions) spécifiques et ciblées au niveau d’un ADN plasmidique double brin. Elle présente de nombreuses applications, notamment les suivantes :
Étude des changements d’activité protéique résultant de la modification de l’ADN ;
Sélection ou criblage de mutations (au niveau de l’ADN, de l’ARN ou de la protéine) présentant une propriété recherchée ;
Ajout ou suppression de sites d’endonucléase de restriction ou de tags.
La mutagenèse dirigée est un processus in vitro qui utilise des amorces oligonucléotidiques spécifiquement conçues pour créer la mutation désirée dans un plasmide. Par le passé, une méthode mise au point par Kunkel en 1985 était largement répandue. Dans cette approche, une souche d’E. coli déficiente en dUTPase et en uracile-ADN glycosylase est utilisée pour produire des copies uracilées de l’ADN d’intérêt. Ces dernières sont ensuite introduites dans une souche sauvage, où elles servent de matrice – en présence d’une amorce portant la mutation désirée – pour la synthèse d’un ADN muté, avant d’être dégradées.
Aujourd’hui, il existe sur le marché un certain nombre de kits qui nécessitent également une modification et/ou des souches d’E. coli spécifiques (par exemple, Phusion Site-Directed Mutagenesis® Kit et le système GeneArt® de Thermo Fisher Scientific). Les méthodes les plus couramment utilisées ne requièrent aucune modification ou souche particulière : les mutations sont incorporées dans le plasmide par PCR inverse à l’aide d’amorces standards, qui peuvent être conçues soit avec un chevauchement (QuikChange®, Agilent), soit avec une orientation dos à dos ( Q5® Site-Directed Mutagenesis Kit ) (figure 1).
Des amorces chevauchantes donnent lieu à un produit qui se recircularise ensuite pour former un plasmide comportant deux coupures simple brin. Malgré la présence de ces dernières, ce produit circulaire peut être utilisé directement pour transformer E. coli, avec une efficacité toutefois inférieure par rapport à un plasmide intact. Les méthodes axées sur des amorces dos à dos offrent non seulement l’avantage de produire des plasmides sans coupures, mais permettent également de générer une quantité bien plus importante du produit désiré, grâce à une amplification exponentielle (figure 2). De plus, comme les amorces ne se chevauchent pas, la taille des délétions n’est limitée que par le plasmide ; et la longueur des insertions, par les contraintes de la synthèse d’oligonucléotides. À l’heure actuelle, en répartissant les bases à ajouter entre les deux amorces, des insertions allant jusqu’à 100 pb peuvent être réalisées en une étape avec cette méthode.
Avant de concevoir les amorces, il est important de déterminer les étapes de mutagenèse à suivre. Nous présentons ici une comparaison de trois kits disponibles sur le marché (figure 3), ainsi qu’une brève description de leurs principales caractéristiques.
So you have finished reading the q5 site directed mutagenesis kit topic article, if you find this article useful, please share it. Thank you very much. See more: q5 site-directed mutagenesis kit (without competent cells), q5® site-directed mutagenesis kit pdf, q5® site-directed mutagenesis kit price, q5 site-directed mutagenesis kit protocol, site-directed mutagenesis kit neb, q5 site-directed mutagenesis troubleshooting, quikchange site-directed mutagenesis kit, agilent site-directed mutagenesis

