You are looking for information, articles, knowledge about the topic nail salons open on sunday near me read_csv encode on Google, you do not find the information you need! Here are the best content compiled and compiled by the https://chewathai27.com/to team, along with other related topics such as: read_csv encode Read_csv, Skiprows pandas, Pandas read_csv, Read CSV Python, Pandas read CSV dtype, Read_csv in R, Pandas read_csv example, Read csv without index pandas
What is encoding in read_csv?
Source from Kaggle character encoding. The Pandas read_csv() function has an argument call encoding that allows you to specify an encoding to use when reading a file. Let’s take a look at an example below: First, we create a DataFrame with some Chinese characters and save it with encoding=’gb2312′ .
What data type does read_csv return?
Read a CSV File
In this case, the Pandas read_csv() function returns a new DataFrame with the data and labels from the file data. csv , which you specified with the first argument. This string can be any valid path, including URLs.
Is read_csv faster than Read_excel?
Importing csv files in Python is 100x faster than Excel files. We can now load these files in 0.63 seconds. That’s nearly 10 times faster! Python loads CSV files 100 times faster than Excel files.
What does CSV in read_csv () stand for?
A comma-separated values (csv) file is returned as two-dimensional data structure with labeled axes. See also DataFrame.to_csv. Write DataFrame to a comma-separated values (csv) file. read_csv. Read a comma-separated values (csv) file into DataFrame.
What is the difference between UTF-8 and ISO 8859 1?
UTF-8 is a multibyte encoding that can represent any Unicode character. ISO 8859-1 is a single-byte encoding that can represent the first 256 Unicode characters. Both encode ASCII exactly the same way.
What encoding does pandas use?
By default, Pandas read_csv() uses a C parser engine for high performance.
How Fast Is pandas read_csv?
…
Reading a CSV with PyArrow.
| CSV parser | Elapsed time | CPU time (user+sys) |
|---|---|---|
| Default C | 13.2 seconds | 13.2 seconds |
| PyArrow | 2.7 seconds | 6.5 seconds |
How does the read_csv function determine column types when reading in a dataset?
When you run read_csv() it prints out a column specification that gives the name and type of each column. That’s an important part of readr, which we’ll come back to in parsing a file. In both cases read_csv() uses the first line of the data for the column names, which is a very common convention.
What is header in PD read_csv?
df.read_csv(‘C:/Users/abc/Desktop/file_name.csv’) header: this allows you to specify which row will be used as column names for your dataframe. Expected an int value or a list of int values. Default value is header=0 , which means the first row of the CSV file will be treated as column names.
Why is openpyxl so slow?
Mike has already provided the solution but here’s the reason for poor performance: the way you’re accessing cells is causing openpyxl to repeatedly parse the original spreadsheet. read-only mode is optimised for row-by-row access.
Is openpyxl faster than pandas?
Step 3: Load with Openpyxl
Still slow but a tiny drop faster than Pandas. Openpyxl Documentation: Memory use is fairly high in comparison with other libraries and applications and is approximately 50 times the original file size.
Is read_excel slow?
read_excel()) is really, really slow, even some with small datasets (<50000 rows), it could take minutes. To speed it up, we are going to convert the Excel files from . xlsx to . csv and use panda.
Does PD read CSV close file?
If you pass it an open file it will keep it open (reading from the current position), if you pass a string then read_csv will open and close the file. In python if you open a file but forget to close it, python will close it for you at the end of the function block (during garbage collection).
How do I save a CSV file without indexing?
In order to export pandas DataFrame to CSV without index (no row indices) use param index=False and to ignore/remove header use header=False param on to_csv() method.
How do I code a CSV file in Python?
- First, open the CSV file for writing ( w mode) by using the open() function.
- Second, create a CSV writer object by calling the writer() function of the csv module.
- Third, write data to CSV file by calling the writerow() or writerows() method of the CSV writer object.
What is utf8 in Python?
UTF-8 is one of the most commonly used encodings, and Python often defaults to using it. UTF stands for “Unicode Transformation Format”, and the ‘8’ means that 8-bit values are used in the encoding.
What is SIG utf8?
“sig” in “utf-8-sig” is the abbreviation of “signature” (i.e. signature utf-8 file). Using utf-8-sig to read a file will treat BOM as file info. instead of a string.
What is Unicode decode error in Python?
The Python “UnicodeDecodeError: ‘ascii’ codec can’t decode byte in position” occurs when we use the ascii codec to decode bytes that were encoded using a different codec. To solve the error, specify the correct encoding, e.g. utf-8 . Here is an example of how the error occurs.
Which argument do you specify with Read_csv to specify a separator character?
read_csv() we have to pass the sep & engine arguments to pandas. read_csv() i.e. Here, sep argument will be used as separator or delimiter. If sep argument is not specified then default engine for parsing ( C Engine) will be used which uses ‘,’ as delimiter.
pandas.read_csv — pandas 1.4.3 documentation
- Article author: pandas.pydata.org
- Reviews from users: 17081
Ratings
- Top rated: 4.5
- Lowest rated: 1
- Summary of article content: Articles about pandas.read_csv — pandas 1.4.3 documentation Encoding to use for UTF when reading/writing (ex. ‘utf-8’). List of Python standard encodings . Changed in version 1.2: When encoding is … …
- Most searched keywords: Whether you are looking for pandas.read_csv — pandas 1.4.3 documentation Encoding to use for UTF when reading/writing (ex. ‘utf-8’). List of Python standard encodings . Changed in version 1.2: When encoding is …
- Table of Contents:

csv – Encoding Error in Panda read_csv – Stack Overflow
- Article author: stackoverflow.com
- Reviews from users: 23231
Ratings
- Top rated: 3.3
- Lowest rated: 1
- Summary of article content: Articles about csv – Encoding Error in Panda read_csv – Stack Overflow Try calling read_csv with encoding=’latin1′ , encoding=’iso-8859-1′ or encoding=’cp1252′ (these are some of the various encodings found on … …
- Most searched keywords: Whether you are looking for csv – Encoding Error in Panda read_csv – Stack Overflow Try calling read_csv with encoding=’latin1′ , encoding=’iso-8859-1′ or encoding=’cp1252′ (these are some of the various encodings found on …
- Table of Contents:
2 Answers
2
Not the answer you’re looking for Browse other questions tagged csv pandas utf-8 or ask your own question

csv – Encoding Error in Panda read_csv – Stack Overflow
- Article author: towardsdatascience.com
- Reviews from users: 31194
Ratings
- Top rated: 4.0
- Lowest rated: 1
- Summary of article content: Articles about csv – Encoding Error in Panda read_csv – Stack Overflow Updating …
- Most searched keywords: Whether you are looking for csv – Encoding Error in Panda read_csv – Stack Overflow Updating
- Table of Contents:
2 Answers
2
Not the answer you’re looking for Browse other questions tagged csv pandas utf-8 or ask your own question

Pandas: How to Read and Write Files – Real Python
- Article author: realpython.com
- Reviews from users: 46906
Ratings
- Top rated: 4.8
- Lowest rated: 1
- Summary of article content: Articles about Pandas: How to Read and Write Files – Real Python Updating …
- Most searched keywords: Whether you are looking for Pandas: How to Read and Write Files – Real Python Updating In this tutorial, you’ll learn about the Pandas IO tools API and how you can use it to read and write files. You’ll use the Pandas read_csv() function to work with CSV files. You’ll also cover similar methods for efficiently working with Excel, CSV, JSON, HTML, SQL, pickle, and big data files.
- Table of Contents:
Installing Pandas
Preparing Data
Using the Pandas read_csv() and to_csv() Functions
Using Pandas to Write and Read Excel Files
Understanding the Pandas IO API
Working With Different File Types
Working With Big Data
Conclusion
Keep reading Real Python by creating a free account or signing in

Pandas: How to Read and Write Files – Real Python
- Article author: towardsdatascience.com
- Reviews from users: 11321
Ratings
- Top rated: 3.6
- Lowest rated: 1
- Summary of article content: Articles about Pandas: How to Read and Write Files – Real Python Updating …
- Most searched keywords: Whether you are looking for Pandas: How to Read and Write Files – Real Python Updating In this tutorial, you’ll learn about the Pandas IO tools API and how you can use it to read and write files. You’ll use the Pandas read_csv() function to work with CSV files. You’ll also cover similar methods for efficiently working with Excel, CSV, JSON, HTML, SQL, pickle, and big data files.
- Table of Contents:
Installing Pandas
Preparing Data
Using the Pandas read_csv() and to_csv() Functions
Using Pandas to Write and Read Excel Files
Understanding the Pandas IO API
Working With Different File Types
Working With Big Data
Conclusion
Keep reading Real Python by creating a free account or signing in

pandas.read_csv — pandas 1.4.3 documentation
- Article author: pandas.pydata.org
- Reviews from users: 22860
Ratings
- Top rated: 4.6
- Lowest rated: 1
- Summary of article content: Articles about pandas.read_csv — pandas 1.4.3 documentation Updating …
- Most searched keywords: Whether you are looking for pandas.read_csv — pandas 1.4.3 documentation Updating
- Table of Contents:

How to read CSV File using Pandas DataFrame.read_csv()
- Article author: net-informations.com
- Reviews from users: 46153
Ratings
- Top rated: 3.2
- Lowest rated: 1
- Summary of article content: Articles about How to read CSV File using Pandas DataFrame.read_csv() DataFrame.read_csv is an important pandas function to read csv files and do operations on it. … pd.read_csv(‘data.csv’, encoding=’utf-8′) … …
- Most searched keywords: Whether you are looking for How to read CSV File using Pandas DataFrame.read_csv() DataFrame.read_csv is an important pandas function to read csv files and do operations on it. … pd.read_csv(‘data.csv’, encoding=’utf-8′) … Importing data from datafile (eg. .csv) is the first step in any data analysis project. DataFrame.read_csv is an important pandas function to read csv files and do operations on it.
- Table of Contents:
Specifying Delimiter
Reading specific Columns only
Read CSV without headers
Skiprows
Use a specific encoding (eg ‘utf-8’ )
Parsing date columns
Specify dType
Multi-character separator
UnicodeDecodeError while read_csv()
Unnamed 0 while read_csv()
Error tokenizing data while read_csv()
FileNotFoundError
MemoryError

Solve Pandas read_csv: UnicodeDecodeError: ‘utf-8’ codec can’t decode byte […] in position […] invalid continuation byte — Roel Peters
- Article author: www.roelpeters.be
- Reviews from users: 20895
Ratings
- Top rated: 4.2
- Lowest rated: 1
- Summary of article content: Articles about Solve Pandas read_csv: UnicodeDecodeError: ‘utf-8’ codec can’t decode byte […] in position […] invalid continuation byte — Roel Peters Okay, so how do I solve it? If you know the encoding of the file, you can simply pass it to the read_csv function, using the encoding parameter. …
- Most searched keywords: Whether you are looking for Solve Pandas read_csv: UnicodeDecodeError: ‘utf-8′ codec can’t decode byte […] in position […] invalid continuation byte — Roel Peters Okay, so how do I solve it? If you know the encoding of the file, you can simply pass it to the read_csv function, using the encoding parameter. Solving the UnicodeDecodeError when using Pandas’ read_csv can be done in multiple ways. In this blog post, I list three.
- Table of Contents:
3 thoughts on “Solve Pandas read_csv UnicodeDecodeError ‘utf-8’ codec can’t decode byte […] in position […] invalid continuation byte”
Related Posts
![Solve Pandas read_csv: UnicodeDecodeError: 'utf-8' codec can't decode byte [...] in position [...] invalid continuation byte — Roel Peters](https://www.roelpeters.be/wp-content/uploads/2021/08/image-1.png)
Solve Pandas read_csv: UnicodeDecodeError: ‘utf-8’ codec can’t decode byte […] in position […] invalid continuation byte — Roel Peters
- Article author: towardsdatascience.com
- Reviews from users: 46082
Ratings
- Top rated: 5.0
- Lowest rated: 1
- Summary of article content: Articles about Solve Pandas read_csv: UnicodeDecodeError: ‘utf-8’ codec can’t decode byte […] in position […] invalid continuation byte — Roel Peters The Pandas read_csv() function has an argument call encoding that allows you to specify an encoding to use when reading a file. …
- Most searched keywords: Whether you are looking for Solve Pandas read_csv: UnicodeDecodeError: ‘utf-8′ codec can’t decode byte […] in position […] invalid continuation byte — Roel Peters The Pandas read_csv() function has an argument call encoding that allows you to specify an encoding to use when reading a file. Solving the UnicodeDecodeError when using Pandas’ read_csv can be done in multiple ways. In this blog post, I list three.
- Table of Contents:
3 thoughts on “Solve Pandas read_csv UnicodeDecodeError ‘utf-8’ codec can’t decode byte […] in position […] invalid continuation byte”
Related Posts
![Solve Pandas read_csv: UnicodeDecodeError: 'utf-8' codec can't decode byte [...] in position [...] invalid continuation byte — Roel Peters](https://www.roelpeters.be/wp-content/uploads/2021/08/image-1.png)
How to Fix – UnicodeDecodeError: invalid start byte – during read_csv in Pandas
- Article author: datascientyst.com
- Reviews from users: 8944
Ratings
- Top rated: 4.7
- Lowest rated: 1
- Summary of article content: Articles about How to Fix – UnicodeDecodeError: invalid start byte – during read_csv in Pandas To use different encoding we can use parameter: encoding : df = pd.read_csv(‘. …
- Most searched keywords: Whether you are looking for How to Fix – UnicodeDecodeError: invalid start byte – during read_csv in Pandas To use different encoding we can use parameter: encoding : df = pd.read_csv(‘.
- Table of Contents:
Getting started
DataFrame Attributes
Import
Export
Access Data
Modify DataFrame
String operation
Special operation
Merge & Concat
Data
Visualization
Challenge
Step 1 UnicodeDecodeError invalid start byte while reading CSV file
Step 2 Solution of UnicodeDecodeError change read_csv encoding
Step 3 Solution of UnicodeDecodeError skip encoding errors with encoding_errors=’ignore’
Step 4 Solution of UnicodeDecodeError fix encoding errors with unicode_escape
Resources
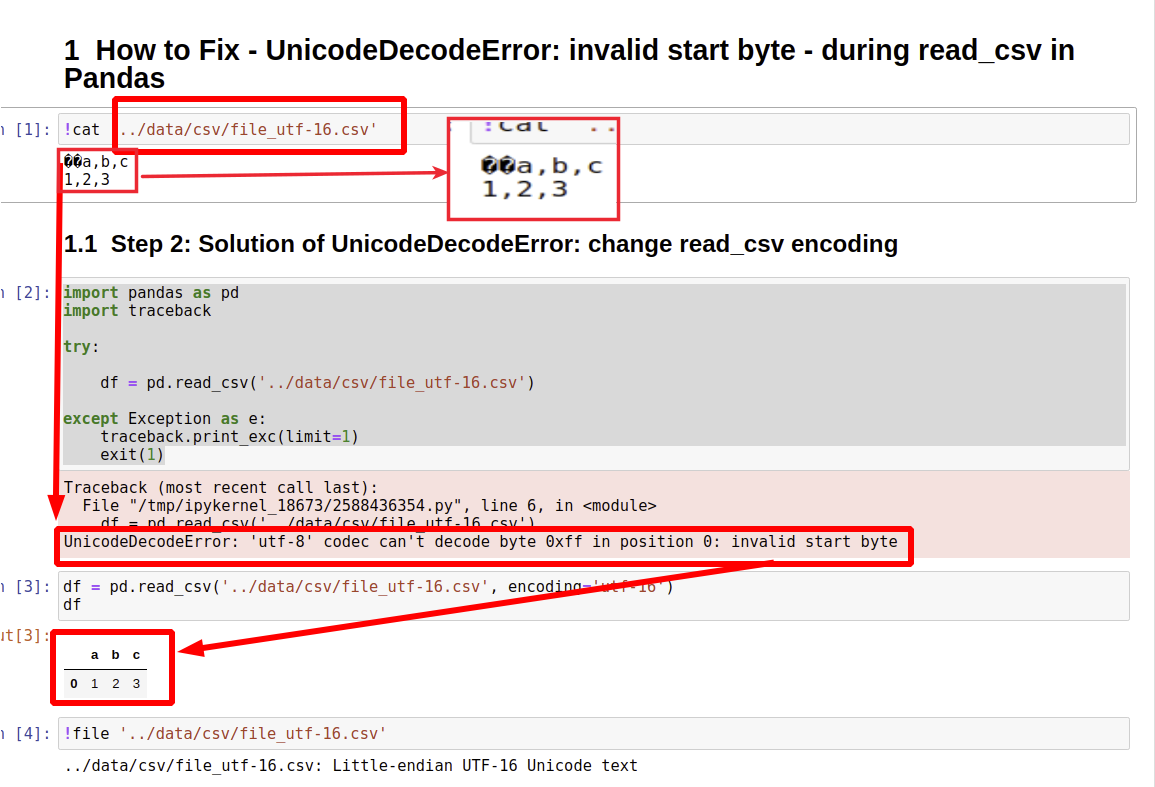
How to Fix – UnicodeDecodeError: invalid start byte – during read_csv in Pandas
- Article author: www.codegrepper.com
- Reviews from users: 17398
Ratings
- Top rated: 4.7
- Lowest rated: 1
- Summary of article content: Articles about How to Fix – UnicodeDecodeError: invalid start byte – during read_csv in Pandas Try calling read_csv with encoding=’latin1′, encoding=’iso-8859-1′ or encoding=’cp1252′. pandas read csv. python by Victorious Vicuña on Mar 20 2020 Comment. …
- Most searched keywords: Whether you are looking for How to Fix – UnicodeDecodeError: invalid start byte – during read_csv in Pandas Try calling read_csv with encoding=’latin1′, encoding=’iso-8859-1′ or encoding=’cp1252′. pandas read csv. python by Victorious Vicuña on Mar 20 2020 Comment.
- Table of Contents:
Getting started
DataFrame Attributes
Import
Export
Access Data
Modify DataFrame
String operation
Special operation
Merge & Concat
Data
Visualization
Challenge
Step 1 UnicodeDecodeError invalid start byte while reading CSV file
Step 2 Solution of UnicodeDecodeError change read_csv encoding
Step 3 Solution of UnicodeDecodeError skip encoding errors with encoding_errors=’ignore’
Step 4 Solution of UnicodeDecodeError fix encoding errors with unicode_escape
Resources

How to resolve a UnicodeDecodeError for a CSV file | Kaggle
- Article author: www.kaggle.com
- Reviews from users: 28002
Ratings
- Top rated: 3.0
- Lowest rated: 1
- Summary of article content: Articles about How to resolve a UnicodeDecodeError for a CSV file | Kaggle Step 1: Try to load the .csv file using pd.read_csv() … Step 3: Now you can manually define the encoding in pd.read_csv(). …
- Most searched keywords: Whether you are looking for How to resolve a UnicodeDecodeError for a CSV file | Kaggle Step 1: Try to load the .csv file using pd.read_csv() … Step 3: Now you can manually define the encoding in pd.read_csv(). Explore and run machine learning code with Kaggle Notebooks | Using data from Demographics of Academy Awards (Oscars) Winners
- Table of Contents:

See more articles in the same category here: Chewathai27.com/to/blog.
csv — pandas 1.4.3 documentation
filepath_or_buffer str, path object or file-like object
Any valid string path is acceptable. The string could be a URL. Valid URL schemes include http, ftp, s3, gs, and file. For file URLs, a host is expected. A local file could be: file://localhost/path/to/table.csv. If you want to pass in a path object, pandas accepts any os.PathLike . By file-like object, we refer to objects with a read() method, such as a file handle (e.g. via builtin open function) or StringIO .
sep str, default ‘,’
Delimiter to use. If sep is None, the C engine cannot automatically detect the separator, but the Python parsing engine can, meaning the latter will be used and automatically detect the separator by Python’s builtin sniffer tool, csv.Sniffer . In addition, separators longer than 1 character and different from ‘\s+’ will be interpreted as regular expressions and will also force the use of the Python parsing engine. Note that regex delimiters are prone to ignoring quoted data. Regex example: ‘\r\t’ .
delimiter str, default None
Alias for sep.
header int, list of int, None, default ‘infer’
Row number(s) to use as the column names, and the start of the data. Default behavior is to infer the column names: if no names are passed the behavior is identical to header=0 and column names are inferred from the first line of the file, if column names are passed explicitly then the behavior is identical to header=None . Explicitly pass header=0 to be able to replace existing names. The header can be a list of integers that specify row locations for a multi-index on the columns e.g. [0,1,3]. Intervening rows that are not specified will be skipped (e.g. 2 in this example is skipped). Note that this parameter ignores commented lines and empty lines if skip_blank_lines=True , so header=0 denotes the first line of data rather than the first line of the file.
names array-like, optional
List of column names to use. If the file contains a header row, then you should explicitly pass header=0 to override the column names. Duplicates in this list are not allowed.
index_col int, str, sequence of int / str, or False, optional, default None
Column(s) to use as the row labels of the DataFrame , either given as string name or column index. If a sequence of int / str is given, a MultiIndex is used. Note: index_col=False can be used to force pandas to not use the first column as the index, e.g. when you have a malformed file with delimiters at the end of each line.
usecols list-like or callable, optional
Return a subset of the columns. If list-like, all elements must either be positional (i.e. integer indices into the document columns) or strings that correspond to column names provided either by the user in names or inferred from the document header row(s). If names are given, the document header row(s) are not taken into account. For example, a valid list-like usecols parameter would be [0, 1, 2] or [‘foo’, ‘bar’, ‘baz’] . Element order is ignored, so usecols=[0, 1] is the same as [1, 0] . To instantiate a DataFrame from data with element order preserved use pd.read_csv(data, usecols=[‘foo’, ‘bar’])[[‘foo’, ‘bar’]] for columns in [‘foo’, ‘bar’] order or pd.read_csv(data, usecols=[‘foo’, ‘bar’])[[‘bar’, ‘foo’]] for [‘bar’, ‘foo’] order. If callable, the callable function will be evaluated against the column names, returning names where the callable function evaluates to True. An example of a valid callable argument would be lambda x: x.upper() in [‘AAA’, ‘BBB’, ‘DDD’] . Using this parameter results in much faster parsing time and lower memory usage.
squeeze bool, default False
If the parsed data only contains one column then return a Series. Deprecated since version 1.4.0: Append .squeeze(“columns”) to the call to read_csv to squeeze the data.
prefix str, optional
Prefix to add to column numbers when no header, e.g. ‘X’ for X0, X1, … Deprecated since version 1.4.0: Use a list comprehension on the DataFrame’s columns after calling read_csv .
mangle_dupe_cols bool, default True
Duplicate columns will be specified as ‘X’, ‘X.1’, …’X.N’, rather than ‘X’…’X’. Passing in False will cause data to be overwritten if there are duplicate names in the columns.
dtype Type name or dict of column -> type, optional
Data type for data or columns. E.g. {‘a’: np.float64, ‘b’: np.int32, ‘c’: ‘Int64’} Use str or object together with suitable na_values settings to preserve and not interpret dtype. If converters are specified, they will be applied INSTEAD of dtype conversion.
engine {‘c’, ‘python’, ‘pyarrow’}, optional
Parser engine to use. The C and pyarrow engines are faster, while the python engine is currently more feature-complete. Multithreading is currently only supported by the pyarrow engine. New in version 1.4.0: The “pyarrow” engine was added as an experimental engine, and some features are unsupported, or may not work correctly, with this engine.
converters dict, optional
Dict of functions for converting values in certain columns. Keys can either be integers or column labels.
true_values list, optional
Values to consider as True.
false_values list, optional
Values to consider as False.
skipinitialspace bool, default False
Skip spaces after delimiter.
skiprows list-like, int or callable, optional
Line numbers to skip (0-indexed) or number of lines to skip (int) at the start of the file. If callable, the callable function will be evaluated against the row indices, returning True if the row should be skipped and False otherwise. An example of a valid callable argument would be lambda x: x in [0, 2] .
skipfooter int, default 0
Number of lines at bottom of file to skip (Unsupported with engine=’c’).
nrows int, optional
Number of rows of file to read. Useful for reading pieces of large files.
na_values scalar, str, list-like, or dict, optional
Additional strings to recognize as NA/NaN. If dict passed, specific per-column NA values. By default the following values are interpreted as NaN: ‘’, ‘#N/A’, ‘#N/A N/A’, ‘#NA’, ‘-1.#IND’, ‘-1.#QNAN’, ‘-NaN’, ‘-nan’, ‘1.#IND’, ‘1.#QNAN’, ‘
’, ‘N/A’, ‘NA’, ‘NULL’, ‘NaN’, ‘n/a’, ‘nan’, ‘null’. keep_default_na bool, default True
Whether or not to include the default NaN values when parsing the data. Depending on whether na_values is passed in, the behavior is as follows: If keep_default_na is True, and na_values are specified, na_values is appended to the default NaN values used for parsing.
If keep_default_na is True, and na_values are not specified, only the default NaN values are used for parsing.
If keep_default_na is False, and na_values are specified, only the NaN values specified na_values are used for parsing.
If keep_default_na is False, and na_values are not specified, no strings will be parsed as NaN. Note that if na_filter is passed in as False, the keep_default_na and na_values parameters will be ignored.
na_filter bool, default True
Detect missing value markers (empty strings and the value of na_values). In data without any NAs, passing na_filter=False can improve the performance of reading a large file.
verbose bool, default False
Indicate number of NA values placed in non-numeric columns.
skip_blank_lines bool, default True
If True, skip over blank lines rather than interpreting as NaN values.
parse_dates bool or list of int or names or list of lists or dict, default False
The behavior is as follows: boolean. If True -> try parsing the index.
list of int or names. e.g. If [1, 2, 3] -> try parsing columns 1, 2, 3 each as a separate date column.
list of lists. e.g. If [[1, 3]] -> combine columns 1 and 3 and parse as a single date column.
dict, e.g. {‘foo’ : [1, 3]} -> parse columns 1, 3 as date and call result ‘foo’ If a column or index cannot be represented as an array of datetimes, say because of an unparsable value or a mixture of timezones, the column or index will be returned unaltered as an object data type. For non-standard datetime parsing, use pd.to_datetime after pd.read_csv . To parse an index or column with a mixture of timezones, specify date_parser to be a partially-applied pandas.to_datetime() with utc=True . See Parsing a CSV with mixed timezones for more. Note: A fast-path exists for iso8601-formatted dates.
infer_datetime_format bool, default False
If True and parse_dates is enabled, pandas will attempt to infer the format of the datetime strings in the columns, and if it can be inferred, switch to a faster method of parsing them. In some cases this can increase the parsing speed by 5-10x.
keep_date_col bool, default False
If True and parse_dates specifies combining multiple columns then keep the original columns.
date_parser function, optional
Function to use for converting a sequence of string columns to an array of datetime instances. The default uses dateutil.parser.parser to do the conversion. Pandas will try to call date_parser in three different ways, advancing to the next if an exception occurs: 1) Pass one or more arrays (as defined by parse_dates ) as arguments; 2) concatenate (row-wise) the string values from the columns defined by parse_dates into a single array and pass that; and 3) call date_parser once for each row using one or more strings (corresponding to the columns defined by parse_dates ) as arguments.
dayfirst bool, default False
DD/MM format dates, international and European format.
cache_dates bool, default True
If True, use a cache of unique, converted dates to apply the datetime conversion. May produce significant speed-up when parsing duplicate date strings, especially ones with timezone offsets. New in version 0.25.0.
iterator bool, default False
Return TextFileReader object for iteration or getting chunks with get_chunk() . Changed in version 1.2: TextFileReader is a context manager.
chunksize int, optional
Return TextFileReader object for iteration. See the IO Tools docs for more information on iterator and chunksize . Changed in version 1.2: TextFileReader is a context manager.
compression str or dict, default ‘infer’
For on-the-fly decompression of on-disk data. If ‘infer’ and ‘%s’ is path-like, then detect compression from the following extensions: ‘.gz’, ‘.bz2’, ‘.zip’, ‘.xz’, or ‘.zst’ (otherwise no compression). If using ‘zip’, the ZIP file must contain only one data file to be read in. Set to None for no decompression. Can also be a dict with key ‘method’ set to one of { ‘zip’ , ‘gzip’ , ‘bz2’ , ‘zstd’ } and other key-value pairs are forwarded to zipfile.ZipFile , gzip.GzipFile , bz2.BZ2File , or zstandard.ZstdDecompressor , respectively. As an example, the following could be passed for Zstandard decompression using a custom compression dictionary: compression={‘method’: ‘zstd’, ‘dict_data’: my_compression_dict} . Changed in version 1.4.0: Zstandard support.
thousands str, optional
Thousands separator.
decimal str, default ‘.’
Character to recognize as decimal point (e.g. use ‘,’ for European data).
lineterminator str (length 1), optional
Character to break file into lines. Only valid with C parser.
quotechar str (length 1), optional
The character used to denote the start and end of a quoted item. Quoted items can include the delimiter and it will be ignored.
quoting int or csv.QUOTE_* instance, default 0
Control field quoting behavior per csv.QUOTE_* constants. Use one of QUOTE_MINIMAL (0), QUOTE_ALL (1), QUOTE_NONNUMERIC (2) or QUOTE_NONE (3).
doublequote bool, default True
When quotechar is specified and quoting is not QUOTE_NONE , indicate whether or not to interpret two consecutive quotechar elements INSIDE a field as a single quotechar element.
escapechar str (length 1), optional
One-character string used to escape other characters.
comment str, optional
Indicates remainder of line should not be parsed. If found at the beginning of a line, the line will be ignored altogether. This parameter must be a single character. Like empty lines (as long as skip_blank_lines=True ), fully commented lines are ignored by the parameter header but not by skiprows . For example, if comment=’#’ , parsing #empty
a,b,c
1,2,3 with header=0 will result in ‘a,b,c’ being treated as the header.
encoding str, optional
Encoding to use for UTF when reading/writing (ex. ‘utf-8’). List of Python standard encodings . Changed in version 1.2: When encoding is None , errors=”replace” is passed to open() . Otherwise, errors=”strict” is passed to open() . This behavior was previously only the case for engine=”python” . Changed in version 1.3.0: encoding_errors is a new argument. encoding has no longer an influence on how encoding errors are handled.
encoding_errors str, optional, default “strict”
How encoding errors are treated. List of possible values . New in version 1.3.0.
dialect str or csv.Dialect, optional
If provided, this parameter will override values (default or not) for the following parameters: delimiter , doublequote , escapechar , skipinitialspace , quotechar , and quoting . If it is necessary to override values, a ParserWarning will be issued. See csv.Dialect documentation for more details.
error_bad_lines bool, optional, default None
Lines with too many fields (e.g. a csv line with too many commas) will by default cause an exception to be raised, and no DataFrame will be returned. If False, then these “bad lines” will be dropped from the DataFrame that is returned. Deprecated since version 1.3.0: The on_bad_lines parameter should be used instead to specify behavior upon encountering a bad line instead.
warn_bad_lines bool, optional, default None
If error_bad_lines is False, and warn_bad_lines is True, a warning for each “bad line” will be output. Deprecated since version 1.3.0: The on_bad_lines parameter should be used instead to specify behavior upon encountering a bad line instead.
on_bad_lines {‘error’, ‘warn’, ‘skip’} or callable, default ‘error’
Specifies what to do upon encountering a bad line (a line with too many fields). Allowed values are : ‘error’, raise an Exception when a bad line is encountered.
‘warn’, raise a warning when a bad line is encountered and skip that line.
‘skip’, skip bad lines without raising or warning when they are encountered. New in version 1.3.0: callable, function with signature (bad_line: list[str]) -> list[str] | None that will process a single bad line. bad_line is a list of strings split by the sep . If the function returns None , the bad line will be ignored. If the function returns a new list of strings with more elements than expected, a ParserWarning will be emitted while dropping extra elements. Only supported when engine=”python” New in version 1.4.0.
delim_whitespace bool, default False
Specifies whether or not whitespace (e.g. ‘ ‘ or ‘ ‘ ) will be used as the sep. Equivalent to setting sep=’\s+’ . If this option is set to True, nothing should be passed in for the delimiter parameter.
low_memory bool, default True
Internally process the file in chunks, resulting in lower memory use while parsing, but possibly mixed type inference. To ensure no mixed types either set False, or specify the type with the dtype parameter. Note that the entire file is read into a single DataFrame regardless, use the chunksize or iterator parameter to return the data in chunks. (Only valid with C parser).
memory_map bool, default False
If a filepath is provided for filepath_or_buffer , map the file object directly onto memory and access the data directly from there. Using this option can improve performance because there is no longer any I/O overhead.
float_precision str, optional
Specifies which converter the C engine should use for floating-point values. The options are None or ‘high’ for the ordinary converter, ‘legacy’ for the original lower precision pandas converter, and ‘round_trip’ for the round-trip converter. Changed in version 1.2.
storage_options dict, optional
Encoding Error in Panda read_csv
This question already has answers here: UnicodeDecodeError when reading CSV file in Pandas with Python (25 answers) Closed 4 years ago .
I’m attempting to read a CSV file into a Dataframe in Pandas. When I try to do that, I get the following error:
UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0x96 in position 55: invalid start byte
This is from code:
import pandas as pd location = r”C:\Users\khtad\Documents\test.csv” df = pd.read_csv(location, header=0, quotechar='”‘)
This is on a Windows 7 Enterprise Service Pack 1 machine and it seems to apply to every CSV file I create. In this particular case the binary from location 55 is 00101001 and location 54 is 01110011, if that matters.
Saving the file as UTF-8 with a text editor doesn’t seem to help, either. Similarly, adding the param “encoding=’utf-8’ doesn’t work, either–it returns the same error.
What is the most likely cause of this error and are there any workarounds other than abandoning the DataFrame construct for the moment and using the csv module to read in the CSV line-by-line?
csv — pandas 1.4.3 documentation
filepath_or_buffer str, path object or file-like object
Any valid string path is acceptable. The string could be a URL. Valid URL schemes include http, ftp, s3, gs, and file. For file URLs, a host is expected. A local file could be: file://localhost/path/to/table.csv. If you want to pass in a path object, pandas accepts any os.PathLike . By file-like object, we refer to objects with a read() method, such as a file handle (e.g. via builtin open function) or StringIO .
sep str, default ‘,’
Delimiter to use. If sep is None, the C engine cannot automatically detect the separator, but the Python parsing engine can, meaning the latter will be used and automatically detect the separator by Python’s builtin sniffer tool, csv.Sniffer . In addition, separators longer than 1 character and different from ‘\s+’ will be interpreted as regular expressions and will also force the use of the Python parsing engine. Note that regex delimiters are prone to ignoring quoted data. Regex example: ‘\r\t’ .
delimiter str, default None
Alias for sep.
header int, list of int, None, default ‘infer’
Row number(s) to use as the column names, and the start of the data. Default behavior is to infer the column names: if no names are passed the behavior is identical to header=0 and column names are inferred from the first line of the file, if column names are passed explicitly then the behavior is identical to header=None . Explicitly pass header=0 to be able to replace existing names. The header can be a list of integers that specify row locations for a multi-index on the columns e.g. [0,1,3]. Intervening rows that are not specified will be skipped (e.g. 2 in this example is skipped). Note that this parameter ignores commented lines and empty lines if skip_blank_lines=True , so header=0 denotes the first line of data rather than the first line of the file.
names array-like, optional
List of column names to use. If the file contains a header row, then you should explicitly pass header=0 to override the column names. Duplicates in this list are not allowed.
index_col int, str, sequence of int / str, or False, optional, default None
Column(s) to use as the row labels of the DataFrame , either given as string name or column index. If a sequence of int / str is given, a MultiIndex is used. Note: index_col=False can be used to force pandas to not use the first column as the index, e.g. when you have a malformed file with delimiters at the end of each line.
usecols list-like or callable, optional
Return a subset of the columns. If list-like, all elements must either be positional (i.e. integer indices into the document columns) or strings that correspond to column names provided either by the user in names or inferred from the document header row(s). If names are given, the document header row(s) are not taken into account. For example, a valid list-like usecols parameter would be [0, 1, 2] or [‘foo’, ‘bar’, ‘baz’] . Element order is ignored, so usecols=[0, 1] is the same as [1, 0] . To instantiate a DataFrame from data with element order preserved use pd.read_csv(data, usecols=[‘foo’, ‘bar’])[[‘foo’, ‘bar’]] for columns in [‘foo’, ‘bar’] order or pd.read_csv(data, usecols=[‘foo’, ‘bar’])[[‘bar’, ‘foo’]] for [‘bar’, ‘foo’] order. If callable, the callable function will be evaluated against the column names, returning names where the callable function evaluates to True. An example of a valid callable argument would be lambda x: x.upper() in [‘AAA’, ‘BBB’, ‘DDD’] . Using this parameter results in much faster parsing time and lower memory usage.
squeeze bool, default False
If the parsed data only contains one column then return a Series. Deprecated since version 1.4.0: Append .squeeze(“columns”) to the call to read_csv to squeeze the data.
prefix str, optional
Prefix to add to column numbers when no header, e.g. ‘X’ for X0, X1, … Deprecated since version 1.4.0: Use a list comprehension on the DataFrame’s columns after calling read_csv .
mangle_dupe_cols bool, default True
Duplicate columns will be specified as ‘X’, ‘X.1’, …’X.N’, rather than ‘X’…’X’. Passing in False will cause data to be overwritten if there are duplicate names in the columns.
dtype Type name or dict of column -> type, optional
Data type for data or columns. E.g. {‘a’: np.float64, ‘b’: np.int32, ‘c’: ‘Int64’} Use str or object together with suitable na_values settings to preserve and not interpret dtype. If converters are specified, they will be applied INSTEAD of dtype conversion.
engine {‘c’, ‘python’, ‘pyarrow’}, optional
Parser engine to use. The C and pyarrow engines are faster, while the python engine is currently more feature-complete. Multithreading is currently only supported by the pyarrow engine. New in version 1.4.0: The “pyarrow” engine was added as an experimental engine, and some features are unsupported, or may not work correctly, with this engine.
converters dict, optional
Dict of functions for converting values in certain columns. Keys can either be integers or column labels.
true_values list, optional
Values to consider as True.
false_values list, optional
Values to consider as False.
skipinitialspace bool, default False
Skip spaces after delimiter.
skiprows list-like, int or callable, optional
Line numbers to skip (0-indexed) or number of lines to skip (int) at the start of the file. If callable, the callable function will be evaluated against the row indices, returning True if the row should be skipped and False otherwise. An example of a valid callable argument would be lambda x: x in [0, 2] .
skipfooter int, default 0
Number of lines at bottom of file to skip (Unsupported with engine=’c’).
nrows int, optional
Number of rows of file to read. Useful for reading pieces of large files.
na_values scalar, str, list-like, or dict, optional
Additional strings to recognize as NA/NaN. If dict passed, specific per-column NA values. By default the following values are interpreted as NaN: ‘’, ‘#N/A’, ‘#N/A N/A’, ‘#NA’, ‘-1.#IND’, ‘-1.#QNAN’, ‘-NaN’, ‘-nan’, ‘1.#IND’, ‘1.#QNAN’, ‘
’, ‘N/A’, ‘NA’, ‘NULL’, ‘NaN’, ‘n/a’, ‘nan’, ‘null’. keep_default_na bool, default True
Whether or not to include the default NaN values when parsing the data. Depending on whether na_values is passed in, the behavior is as follows: If keep_default_na is True, and na_values are specified, na_values is appended to the default NaN values used for parsing.
If keep_default_na is True, and na_values are not specified, only the default NaN values are used for parsing.
If keep_default_na is False, and na_values are specified, only the NaN values specified na_values are used for parsing.
If keep_default_na is False, and na_values are not specified, no strings will be parsed as NaN. Note that if na_filter is passed in as False, the keep_default_na and na_values parameters will be ignored.
na_filter bool, default True
Detect missing value markers (empty strings and the value of na_values). In data without any NAs, passing na_filter=False can improve the performance of reading a large file.
verbose bool, default False
Indicate number of NA values placed in non-numeric columns.
skip_blank_lines bool, default True
If True, skip over blank lines rather than interpreting as NaN values.
parse_dates bool or list of int or names or list of lists or dict, default False
The behavior is as follows: boolean. If True -> try parsing the index.
list of int or names. e.g. If [1, 2, 3] -> try parsing columns 1, 2, 3 each as a separate date column.
list of lists. e.g. If [[1, 3]] -> combine columns 1 and 3 and parse as a single date column.
dict, e.g. {‘foo’ : [1, 3]} -> parse columns 1, 3 as date and call result ‘foo’ If a column or index cannot be represented as an array of datetimes, say because of an unparsable value or a mixture of timezones, the column or index will be returned unaltered as an object data type. For non-standard datetime parsing, use pd.to_datetime after pd.read_csv . To parse an index or column with a mixture of timezones, specify date_parser to be a partially-applied pandas.to_datetime() with utc=True . See Parsing a CSV with mixed timezones for more. Note: A fast-path exists for iso8601-formatted dates.
infer_datetime_format bool, default False
If True and parse_dates is enabled, pandas will attempt to infer the format of the datetime strings in the columns, and if it can be inferred, switch to a faster method of parsing them. In some cases this can increase the parsing speed by 5-10x.
keep_date_col bool, default False
If True and parse_dates specifies combining multiple columns then keep the original columns.
date_parser function, optional
Function to use for converting a sequence of string columns to an array of datetime instances. The default uses dateutil.parser.parser to do the conversion. Pandas will try to call date_parser in three different ways, advancing to the next if an exception occurs: 1) Pass one or more arrays (as defined by parse_dates ) as arguments; 2) concatenate (row-wise) the string values from the columns defined by parse_dates into a single array and pass that; and 3) call date_parser once for each row using one or more strings (corresponding to the columns defined by parse_dates ) as arguments.
dayfirst bool, default False
DD/MM format dates, international and European format.
cache_dates bool, default True
If True, use a cache of unique, converted dates to apply the datetime conversion. May produce significant speed-up when parsing duplicate date strings, especially ones with timezone offsets. New in version 0.25.0.
iterator bool, default False
Return TextFileReader object for iteration or getting chunks with get_chunk() . Changed in version 1.2: TextFileReader is a context manager.
chunksize int, optional
Return TextFileReader object for iteration. See the IO Tools docs for more information on iterator and chunksize . Changed in version 1.2: TextFileReader is a context manager.
compression str or dict, default ‘infer’
For on-the-fly decompression of on-disk data. If ‘infer’ and ‘%s’ is path-like, then detect compression from the following extensions: ‘.gz’, ‘.bz2’, ‘.zip’, ‘.xz’, or ‘.zst’ (otherwise no compression). If using ‘zip’, the ZIP file must contain only one data file to be read in. Set to None for no decompression. Can also be a dict with key ‘method’ set to one of { ‘zip’ , ‘gzip’ , ‘bz2’ , ‘zstd’ } and other key-value pairs are forwarded to zipfile.ZipFile , gzip.GzipFile , bz2.BZ2File , or zstandard.ZstdDecompressor , respectively. As an example, the following could be passed for Zstandard decompression using a custom compression dictionary: compression={‘method’: ‘zstd’, ‘dict_data’: my_compression_dict} . Changed in version 1.4.0: Zstandard support.
thousands str, optional
Thousands separator.
decimal str, default ‘.’
Character to recognize as decimal point (e.g. use ‘,’ for European data).
lineterminator str (length 1), optional
Character to break file into lines. Only valid with C parser.
quotechar str (length 1), optional
The character used to denote the start and end of a quoted item. Quoted items can include the delimiter and it will be ignored.
quoting int or csv.QUOTE_* instance, default 0
Control field quoting behavior per csv.QUOTE_* constants. Use one of QUOTE_MINIMAL (0), QUOTE_ALL (1), QUOTE_NONNUMERIC (2) or QUOTE_NONE (3).
doublequote bool, default True
When quotechar is specified and quoting is not QUOTE_NONE , indicate whether or not to interpret two consecutive quotechar elements INSIDE a field as a single quotechar element.
escapechar str (length 1), optional
One-character string used to escape other characters.
comment str, optional
Indicates remainder of line should not be parsed. If found at the beginning of a line, the line will be ignored altogether. This parameter must be a single character. Like empty lines (as long as skip_blank_lines=True ), fully commented lines are ignored by the parameter header but not by skiprows . For example, if comment=’#’ , parsing #empty
a,b,c
1,2,3 with header=0 will result in ‘a,b,c’ being treated as the header.
encoding str, optional
Encoding to use for UTF when reading/writing (ex. ‘utf-8’). List of Python standard encodings . Changed in version 1.2: When encoding is None , errors=”replace” is passed to open() . Otherwise, errors=”strict” is passed to open() . This behavior was previously only the case for engine=”python” . Changed in version 1.3.0: encoding_errors is a new argument. encoding has no longer an influence on how encoding errors are handled.
encoding_errors str, optional, default “strict”
How encoding errors are treated. List of possible values . New in version 1.3.0.
dialect str or csv.Dialect, optional
If provided, this parameter will override values (default or not) for the following parameters: delimiter , doublequote , escapechar , skipinitialspace , quotechar , and quoting . If it is necessary to override values, a ParserWarning will be issued. See csv.Dialect documentation for more details.
error_bad_lines bool, optional, default None
Lines with too many fields (e.g. a csv line with too many commas) will by default cause an exception to be raised, and no DataFrame will be returned. If False, then these “bad lines” will be dropped from the DataFrame that is returned. Deprecated since version 1.3.0: The on_bad_lines parameter should be used instead to specify behavior upon encountering a bad line instead.
warn_bad_lines bool, optional, default None
If error_bad_lines is False, and warn_bad_lines is True, a warning for each “bad line” will be output. Deprecated since version 1.3.0: The on_bad_lines parameter should be used instead to specify behavior upon encountering a bad line instead.
on_bad_lines {‘error’, ‘warn’, ‘skip’} or callable, default ‘error’
Specifies what to do upon encountering a bad line (a line with too many fields). Allowed values are : ‘error’, raise an Exception when a bad line is encountered.
‘warn’, raise a warning when a bad line is encountered and skip that line.
‘skip’, skip bad lines without raising or warning when they are encountered. New in version 1.3.0: callable, function with signature (bad_line: list[str]) -> list[str] | None that will process a single bad line. bad_line is a list of strings split by the sep . If the function returns None , the bad line will be ignored. If the function returns a new list of strings with more elements than expected, a ParserWarning will be emitted while dropping extra elements. Only supported when engine=”python” New in version 1.4.0.
delim_whitespace bool, default False
Specifies whether or not whitespace (e.g. ‘ ‘ or ‘ ‘ ) will be used as the sep. Equivalent to setting sep=’\s+’ . If this option is set to True, nothing should be passed in for the delimiter parameter.
low_memory bool, default True
Internally process the file in chunks, resulting in lower memory use while parsing, but possibly mixed type inference. To ensure no mixed types either set False, or specify the type with the dtype parameter. Note that the entire file is read into a single DataFrame regardless, use the chunksize or iterator parameter to return the data in chunks. (Only valid with C parser).
memory_map bool, default False
If a filepath is provided for filepath_or_buffer , map the file object directly onto memory and access the data directly from there. Using this option can improve performance because there is no longer any I/O overhead.
float_precision str, optional
Specifies which converter the C engine should use for floating-point values. The options are None or ‘high’ for the ordinary converter, ‘legacy’ for the original lower precision pandas converter, and ‘round_trip’ for the round-trip converter. Changed in version 1.2.
storage_options dict, optional
So you have finished reading the read_csv encode topic article, if you find this article useful, please share it. Thank you very much. See more: Read_csv, Skiprows pandas, Pandas read_csv, Read CSV Python, Pandas read CSV dtype, Read_csv in R, Pandas read_csv example, Read csv without index pandas

